- Activities
- QA4EO-RUT
- Data Resources
Data Resources
Overview
Data Resources
For the implementation of the RUT tool, the author has been working in partnership with Brockmann Consult. The RUT tool is integrated as part of the Sentinels Toolbox. The code is fully accessible in the software repository located in:
- Sentinel-2 Radiometric Uncertainty Tool repository
- Latest installers for SNAP and the Sentinel Toolboxes can be found here This code works as a plug-in that is embedded as part of the Sentinel Toolbox. It creates an operator so that when the user opens a product image, the operator is called and automatically generates the uncertainty image. In the image below, it is shown an example of where the RUT tool is allocated within the Sentinel Toolbox. The main features are:
- SNAP Python libraries (snappy) for product readout.
- General tool design to accommodate other sensors
- Maximisation of product info extraction (e.g. noise coefficients) makes it robust against re-processing, contingencies…
The following sub-sections define the algorithms and processor.
RUT v1 class S2RutOp description
- Step 1: Initialize function
This function defines the product reading, the setup of the uncertainty product output and the global parameters extraction. Specific tasks:
The source product is read (source_product) and verified as a valid Sentinel-2 product.
Parsing Sentinel-2 metadata at three levels: product_meta, datastrip_meta and granule_meta.
Extraction of global metadata parameters: distance to sun (u_sun),reflectance quantification (quant) and mean sun zenith angle (tecta). Note: next versions of the tool to include the per-pixel sun zenith angle.
Extraction of user parameters: selected bands (toa_band_names), coverage factor (k) and uncertainty contributors flags (unc_select).
Setup of the uncertainty product including L1C scene_width, scene_height and geolocation: rut_product = snappy.Product(self.source_product.getName() + '_rut', 'S2_RUT', scene_width, scene_height)
Add each one of the selected bands (unc_toa_band) in the uncertainty product with the correct name and Nodata value.
- Step 2: computeTile function
This function selects the specific band in a tile, extracts the required parameters and sets the uncertainty samples after calling the S2RutAlgo operator. Specific tasks:
Selects the source_band and toa_band_id.
Extracts the metadata parameters for the specific band: absolute calibration coefficient (a), sun irradiance (e_sun), noise model coefficients (alpha and beta), and estimated diffuser degradation (u_diff_temp).
Extracts the L1C samples toa_samples
Call the S2RutAlgo for the per-pixel uncertainty calculation unc = self.rut_algo.unc_calculation(np.array(toa_samples, dtype=np.uint16), toa_band_id)
Set the samples in the destination tile
RUT v1 class S2RutAlgo description
- Step 1: Initial check
This step identifies No data, cloud, defective or saturated pixels.
RUTv1 does not include this step. This is to be implemented in next versions of the RUT
- Step 2: Undo reflectance conversion
This step converts the L1C reflectance in L1B CN counts.
cn = (self.a * self.e_sun * self.u_sun * math.cos(math.radians(self.tecta)) / ( math.pi * self.quant)) * band_data.
Warning raised if tecta > 70°. Conversion error can be > 5%
- Step 3: Orthorectification process
This step evaluates the impact of the uncertainty propagation through the resampling process.
RUTv1 does not include this step. This is to be implemented in next versions of the RUT
- Step 4: L1B uncertainty contributors: raw and dark signal
NOTE: Each the following uncertainty contributors are pre-checked and included if unc_select == True
Noise in % units
u_noise = 100 * np.sqrt(self.alpha ** 2 + self.beta * cn) / cn
Out-of field straylight - systematic in radiance units
u_stray_sys = 0.3 * rad_conf.Lref[band_id] / 100
Out-of field straylight - random in % units
u_stray_rand = rad_conf.u_stray_rand_all[band_id]
Crosstalk in radiance units
u_xtalk = rad_conf.u_xtalk_all[band_id]
Instrument quantisation. Defined in _init_ function.
self.u_ADC = 0.5
Predefined DS stability in LSB units
u_DS = rad_conf.u_DS_all[band_id]
- Step 5: L1B uncertainty contributors: gamma correction
Predefined relative gains residual in % units
u_gamma = 0.4
- Step 6: L1C uncertainty contributors: absolute calibration coefficient
Pre-flight diffuser reflectance knowledge in % units
u_diff_abs = rad_conf.u_diff_absarray[band_id]
u_diff_temp calculated in computeTile function(see above)
Cosine diffuser knowledge. Defined in _init_ function.
self.u_diff_cos = 0.4
Diffuser straylight residual correction. Defined in _init_ function.
self.u_diff_k = 0.3
- Step 7: L1C uncertainty contributors: reflectance conversion
Impact of the reflectance factor digitisation in % units
u_ref_quant = 100 * (0.5 / math.sqrt(3)) / band_data
- Step 8: Combine uncertainty contributors
Calculation of the terms u'DS and u'ADC
u_adc = (100 * self.u_ADC / math.sqrt(3)) / cn ; u_ds = (100 * u_DS) / cn
Calculation of the straylight term and conversion of the crosstalk into % units
u_stray = np.sqrt(u_stray_rand ** 2 + ((100 * self.a * u_xtalk) / cn) ** 2)
Calculation of the diffuser uncertainty
u_diff = math.sqrt(u_diff_abs ** 2 + self.u_diff_cos ** 2 + self.u_diff_k ** 2)
Calculation of the standard uncertainty
u_1sigma = np.sqrt(u_ref_quant ** 2 + u_gamma ** 2 + u_stray ** 2 + u_diff ** 2 + u_noise ** 2 + u_adc ** 2 + u_ds ** 2)
Calculation of the expanded uncertainty in % units. Conversion of the Out-of-Field Straylight - systematic in % units. Resulting uncertainty values multiplied by 10
u_expand = 10 * (self.u_diff_temp + ((100 * self.a * u_stray_sys) / cn) + self.k * u_1sigma)
Clipping all the values to range [0,250] for uncertainty image UINT8 type
u_ref = np.uint8(np.clip(u_expand, 0, 250))
- Step 9: Append uncertainty information to the metadata
This step integrates relevant information (e.g. mean pixel uncertainty) in the uncertainty product metadata.
RUTv1 does not include this step. This is to be implemented in next versions of the RUT
Processor Documentation
The S2-RUT processor can be invoked in the Sentinels Toolbox from the menu by selecting Optical➡ Preprocessing➡Sentinel-2 Radiometric Uncertainty Tool. On the command line processor is available by means of the Graph Processing Tool gpt which is located in the Sentinel Toolbox bin directory. Typing gpt S2Rut -h displays further information.
Selecting the Sentinel-2 Radiometric Uncertainty Tool command from the Sentinel Toolbox menu opens up the dialog in the figure below:
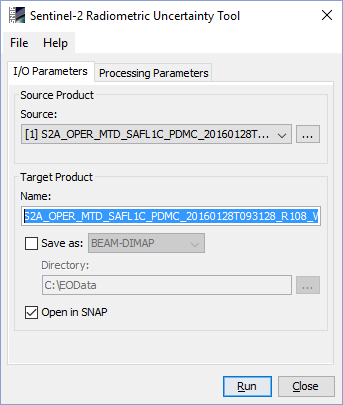
In the "Source product" area, the user specifies the source product. The combo box presents a list of all products opened in the Sentinel Toolbox. The user may select one of these or, by clicking on the button next to the combo box, choose a product from the file system. The selected product must be of type S2_MSI_Level-1C.
In the "Target product" area, the user can specify a name for the generated product. The tool will automatically assign a name by adding the extension "rut" to the Sentinel-2 L1C product selected. The user can also specify where the target product should be saved to the file system. The combo box presents a list of available file formats. The text field or the button next to it allow to specify a target directory. Finally, the option, "Open in SNAP" specifies whether the target product should be opened in the Sentinel Toolbox. When the target product is not saved, it is opened in the Sentinel Toolbox automatically.
The tool incorporates a secondary tab named "Processing Parameters". The figure below presents a screen-shot as it appears in the Sentinels Toolbox.

The option "Coverage factor" permits specifying the k parameter that assigns a probability coverage to the uncertainty evaluation — k=1 approximates 68.27% probability. Another options permits the selection of "Band names" for which the uncertainty shall be computed. Finally, the tab includes a selection list with the uncertainty contributions included in the RUT tool. This permits the user the selection and deselection of specific contributions in order to perform sensitivity analysis, separation of random/systematic contributions…